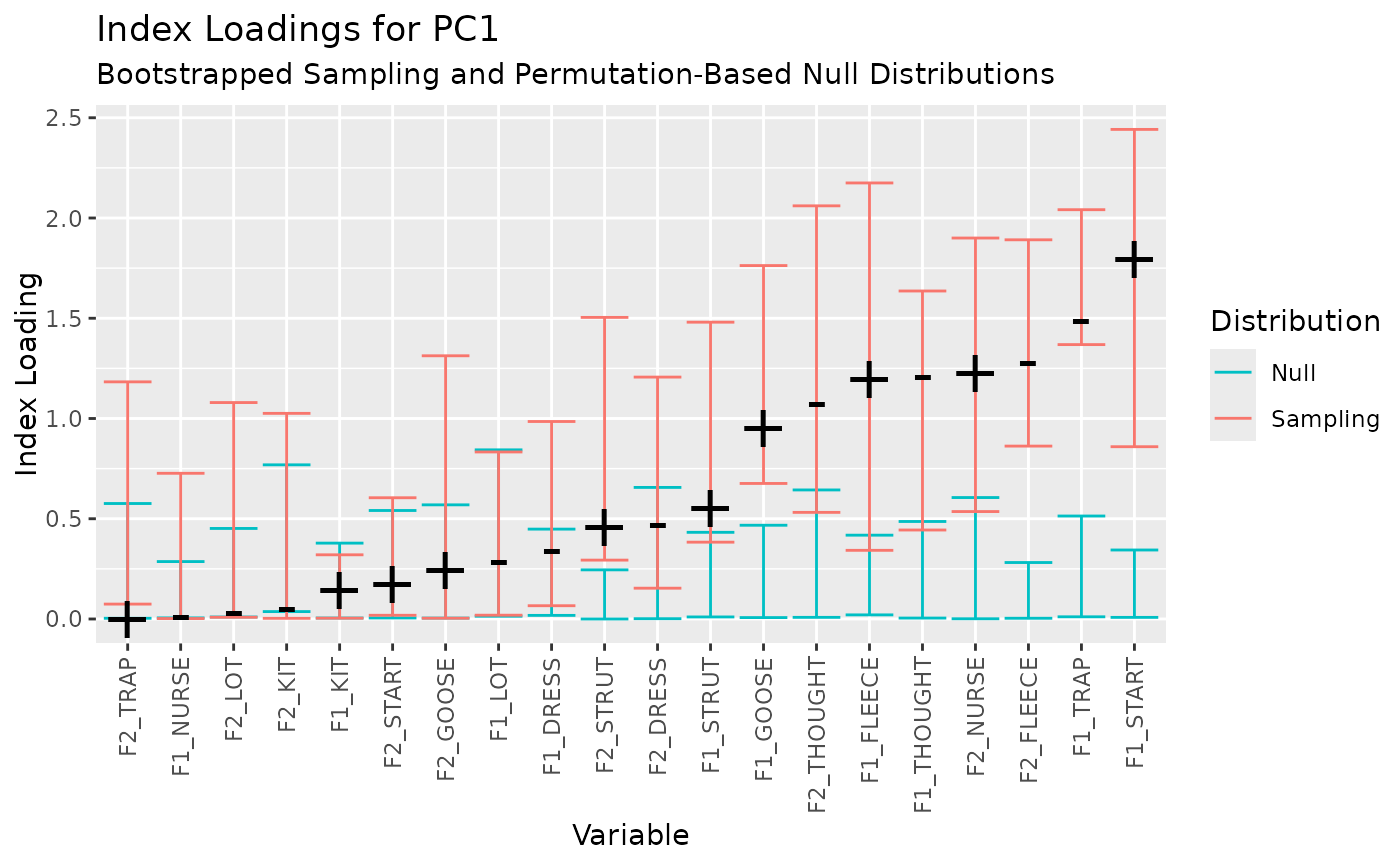
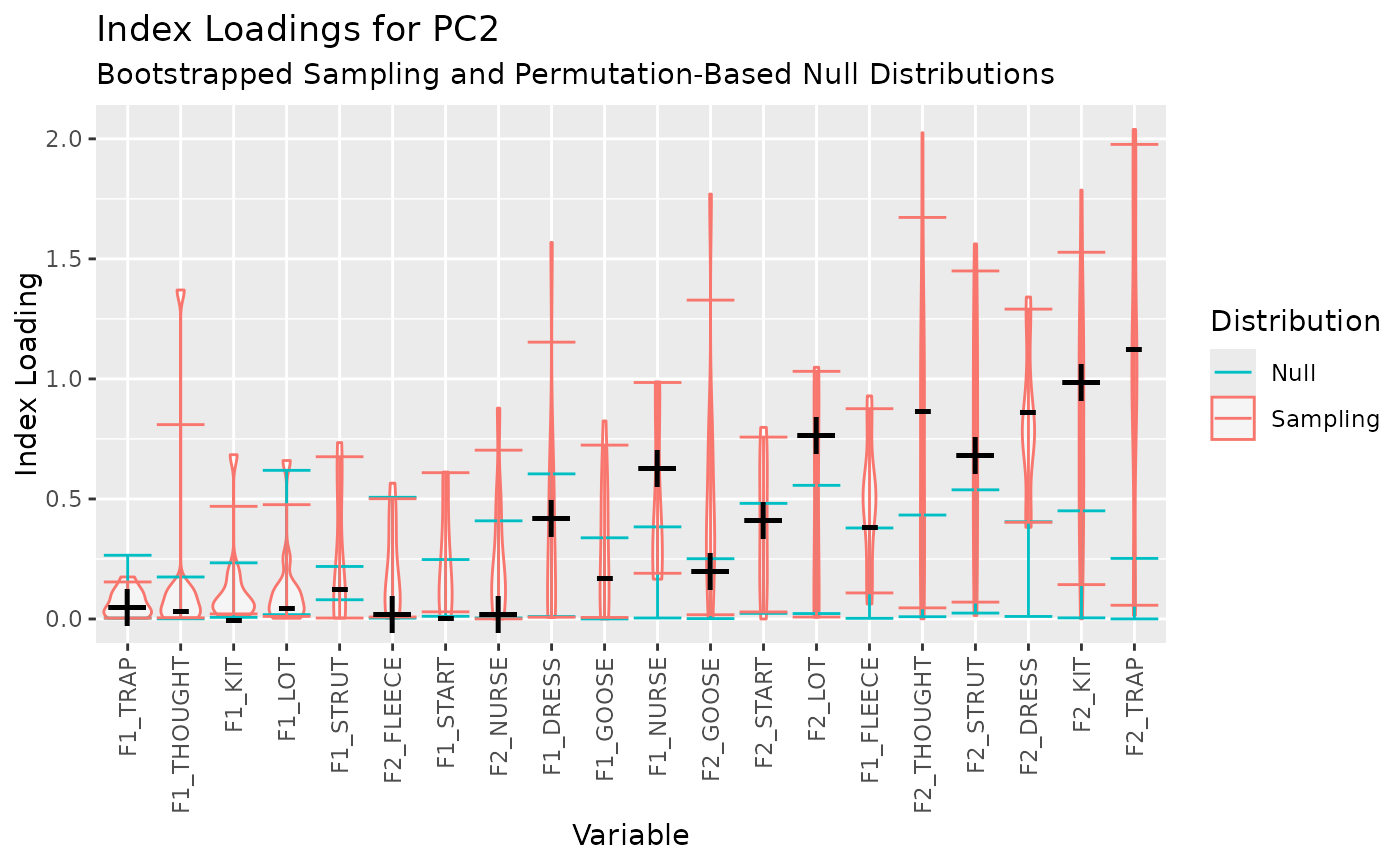
Index loadings (Vieira 2012) are presented with confidence intervals on the sampling distribution generated by bootstrapping and a null distribution generated by permutation.
Usage
plot_loadings(
pca_test,
pc_no = 1,
violin = FALSE,
filter_boots = FALSE,
quantile_threshold = 0.25
)Arguments
- pca_test
an object of class pca_test_results generated by
pca_test.- pc_no
An integer indicating which PC to plot.
- violin
If TRUE, violin plots are added for the confidence intervals of the sampling distribution.
- filter_boots
if TRUE, only bootstrap iterations in which the variable with the highest median loading is above
quantile_threshold.- quantile_threshold
a real value between 0 and 1. Use this to change the threshold used for filtering bootstrap iterations. The default is 0.25.
Details
If PCs are unstable, there is an option (filter_boots) to take only the
bootstrap iterations in which the variable with the highest median loading
across all iterations is above quantile_threshold (default: 0.25). This
helps to reveal reliable connections of this variable with other variables in
the data set.